First chromosome scale genome of Acrocomia aculeata
First chromosome scale genome of Acrocomia aculeata
Scaketti, M.; Garcia, C. B.; Araujo, J. V. d. S. R.; de Carvalho, I. A. S.; Marroquin, J. A. M.; Riano-Pachon, D. M.; Neto, D. D.; Labate, C. A.; Vicentini, R.; Pinheiro, J. B.; Vianna, S.; Colombo, C.; Zucchi, M. I.
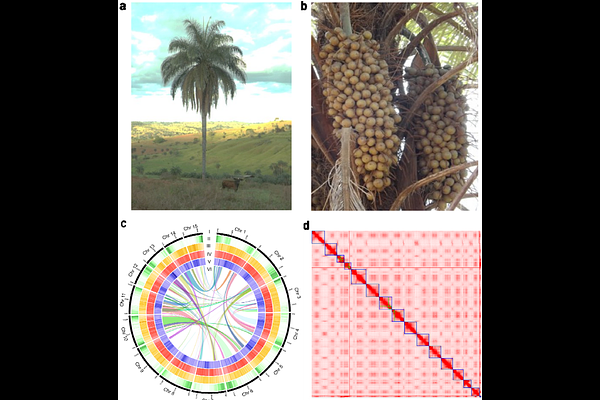
AbstractThe Arecaceae family comprises economically, and ecologically significant palm species widely distributed across tropical and subtropical regions. Among them, Acrocomia aculeata, commonly known as Macauba, has gained attention due to its high oil yield, environmental adaptability, and potential applications in sustainable agriculture and bioenergy production. This study presents the first chromosome scale genome assembly of A. aculeata and the first reference genome of the genus, using Oxford Nanopore Technologies sequencing, PacBio HiFi and Hi-C proximity ligation data. The genome covers 1.94 Gbp in the 15 pseudochromosomes, with N50 of 143.43 Mbp, achieving a base quality QV of 76.4 and a k-mer completeness of 90.58% of PacBio HiFi data. Interspersed repetitive regions represents 77.36% of the total genome, with long terminal repeat (LTRs) accounting for 54.98%. A total of 28.204 protein-coding genes were predicted. The genome was validated with BUSCO analysis, achieving a high completeness of 98.3% for predicted proteins. The first chromosome scale genome of Macauba can provide new insights and breakthroughs for studies of conservation genomics and plant breeding programs.