BioMiner: A Multi-modal System for Automated Mining of Protein-Ligand Bioactivity Data from Literature
BioMiner: A Multi-modal System for Automated Mining of Protein-Ligand Bioactivity Data from Literature
Yan, J.; Zhu, J.; Yang, Y.; Liu, Q.; Zhang, K.; Zhang, Z.; Liu, X.; Zhang, B.; Gao, K.; Xiao, J.; Chen, E.
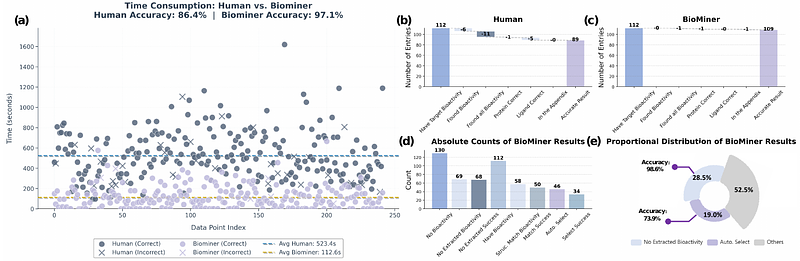
AbstractProtein-ligand bioactivity data published in literature are essential for drug discovery, yet manual curation struggles to keep pace with rapidly growing literature. Automated bioactivity extraction is challenging due to the multi-modal distribution of information (text, tables, figures, structures) and the complexity of chemical representations (e.g., Markush structures). Furthermore, the lack of standardized benchmarks impedes the evaluation and development of extraction methods. In this work, we introduce BioMiner, a multi-modal system designed to automatically extract protein-ligand bioactivity data from thousands to potentially millions of publications. BioMiner employs a modular, agent-based architecture, leveraging a synergistic combination of multi-modal large language models, domain-specific models, and domain tools to navigate this complex extraction task. To address the benchmark gap and support method development, we establish BioVista, a comprehensive benchmark comprising 16,457 bioactivity entries and 8,735 chemical structures curated from 500 publications. On BioVista, BioMiner validates its extraction ability and provides a quantitative baseline, achieving F1 scores of 0.22, 0.45, and 0.53 for bioactivity triplets, chemical structures, and bioactivity measurement with high throughput (14s/paper on 8 V100 GPUs). We further demonstrate BioMiner\'s practical utility via three applications: (1) extracting 67,953 data from 11,683 papers to build a pre-training database that improves downstream models performance by 3.1%; (2) enabling a human-in-the-loop workflow that doubles the number of high-quality NLRP3 bioactivity data, helping 38.6% improvement over 28 QSAR models and identification of 16 hit candidates with novel scaffolds; and (3) accelerating the annotation of the protein-ligand structures in PoseBusters benchmark with reported bioactivity, achieving a 5-fold speed increase and 10% accuracy improvement over manual methods. BioMiner and BioVista provide a scalable extraction methodology and a rigorous benchmark, paving the way to unlock vast amounts of previously inaccessible bioactivity data and accelerate data-driven drug discovery.