Divergence between transcriptomes and chromatin accessibility during differentiation from a bipotential progenitor cell population to erythroblasts and megakaryocytes
Divergence between transcriptomes and chromatin accessibility during differentiation from a bipotential progenitor cell population to erythroblasts and megakaryocytes
Mishra, T.; Giardine, B. M.; Morrissey, C. S.; Keller, C. A.; Heuston, E. F.; Anderson, S. M.; Paralkar, V. R.; Pimkin, M.; Weiss, M. J.; Bodine, D. M.; Hardison, R. C.
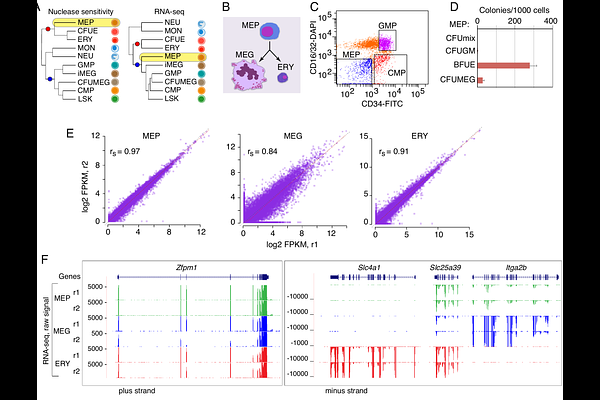
AbstractChanges in gene expression drive differentiation along distinct cell lineages, and these shifts in gene expression are associated with alterations in chromatin accessibility and modifications reflecting activation or repression. We used deep sequencing of polyA+ RNA to map the transcriptomes of the megakaryocyte-erythroid progenitor (MEP) and cells of its two daughter lineages, erythroblasts (ERY) and megakaryocytes (MEG) in mice to reveal insights into differentiation. Transcriptome comparisons revealed that MEPs already expressed much of the MEG program while continuing to express genes associated with parallel myeloid lineages. By contrast, ERY underwent an extensive program of gene induction along with repression of pan-hematopoietic and MEG genes. Maps of transcription factor (TF) occupancy also indicated distinct modes of regulation for the MEG and ERY programs, with MEG genes preferentially occupied by hematopoietic TFs in multipotent progenitors and continued occupancy post-commitment, in contrast to erythroid genes that were primarily occupied in committed ERY. Previous work had indicated a surprising discordance in the clustering of MEP with other hematopoietic cell types by RNA-seq versus chromatin states. We combined the differential expression data with chromatin accessibility across blood cell types to identify trends that contribute to this discordance. Specifically, candidate cis-regulatory elements (cCREs) in some ERY-specific genes were precociously actuated in the bipotential cell populations, and some other genes were expressed in both the MEP population and MEG but their cCREs have less chromatin accessibility in MEP. This discordance in cell type clustering by different modalities of functional genomics may reflect the different contributions of subpopulations in the MEP to the different modalities measured.