decemedip: hierarchical Bayesian modeling for cell type deconvolution of immunoprecipitation-based DNA methylomes
decemedip: hierarchical Bayesian modeling for cell type deconvolution of immunoprecipitation-based DNA methylomes
Shen, N.; Zhang, Z.; Baca, S.; Korthauer, K.
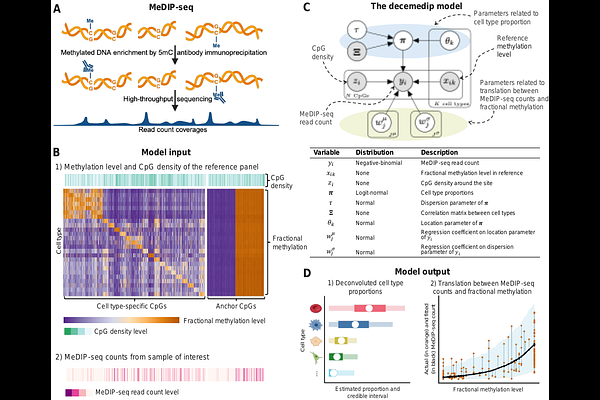
AbstractMeDIP-seq is an enrichment-based DNA methylation profiling technique that measures the abundance of methylated DNA. While this technique offers efficiency advantages over direct methylation profiling, it does not provide absolute quantification of DNA methylation necessary for cell type deconvolution. We introduce decemedip, a Bayesian hierarchical model for cell type deconvolution of methylated sequencing data that leverages reference atlases derived from direct methylation profiling. We demonstrate its accuracy and robustness through simulation studies and validation on cross-platform measurements, and highlight its utility in identifying tissue-specific and cancer-associated methylation signatures using MeDIP-seq profiling of patient-derived xenografts and cell-free DNA. decemedip is available at \\url{https://github.com/nshen7/decemedip}.