Dynamic Contrastive Learning with Pretrained Deep Language Model Enhances Metagenome Binning for Contigs
Dynamic Contrastive Learning with Pretrained Deep Language Model Enhances Metagenome Binning for Contigs
Zou, B.; Zhang, Z.; Tao, R.; Gu, N.; Zhang, L.
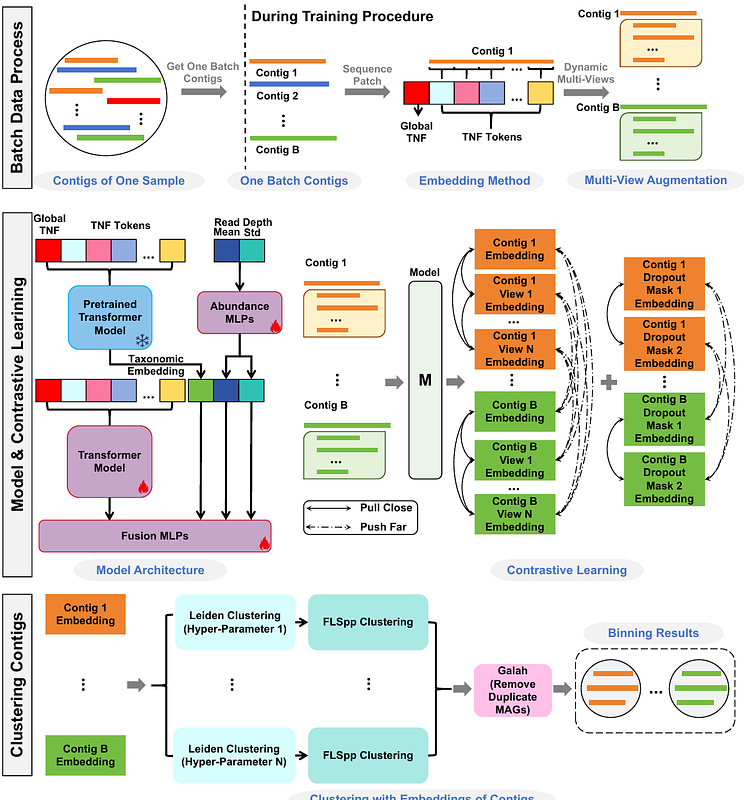
AbstractThe recovery of microbial genomes from large metagenomic datasets is a critical step for identifying uncultivated microbial populations and explaining their functional roles. This process requires metagenomic binning, which involves clustering assembled contigs into metagenome-assembled genomes (MAGs). However, existing computational binning methods often fail to leverage contigs\' sequence context effectively. Most of them cannot incorporate taxonomic knowledge from external databases, and insufficient training data frequently limits the deep learning-based binning methods. To address these challenges, we propose CompleteBin, an innovative pretrained deep language model enhanced by dynamic contrastive learning. CompleteBin trains a pretrained deep language model using both long and short contigs and clusters the contigs based on their embeddings generated by the language model. This approach uniquely integrates the sequence context of contigs, incorporates taxonomic knowledge from external databases, and leverages unlimited data during training to significantly improve binning accuracy. Compared to state-of-the-art binning tools such as CONCOCT, MetaBAT2, SemiBin2, and COMEBin, CompleteBin achieves higher performance across both simulated datasets (CAMI I and II) and diverse real-world datasets, including ocean, plant, freshwater, and human fecal samples. On average, CompleteBin achieves a 39.7% improvement on simulated datasets and a 58.9% improvement on real-world datasets in terms of the number of near-complete MAGs when compared to the best-performing results of other binning methods. These results highlight CompleteBin\'s robust capability to recover high-quality MAGs from long and short contigs, establishing it as a powerful tool for advancing metagenomic research. CompleteBin is open-source and available at https://github.com/zoubohao/CompleteBin.