The structure, folding kinetics and dynamics of long poly(UG) RNA
The structure, folding kinetics and dynamics of long poly(UG) RNA
Petersen, R. J.; Vivek, R.; Tonelli, M.; Roschdi, S.; Butcher, S. E.
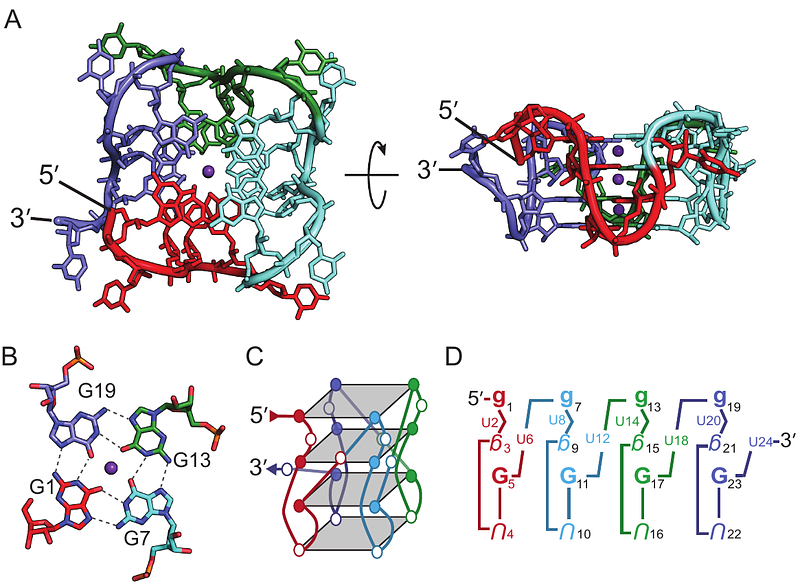
AbstractLong poly(UG) or \"pUG\" dinucleotide repeats are abundant in eukaryotic transcriptomes. Thousands of human genes have pUGs longer than 24 repeats, including the cancer-associated lncRNA NEAT1. In C. elegans, enzymatic addition of long pUGs to RNA 3\' ends (pUG tails) marks RNAs as vectors of gene silencing. Gene silencing requires at least one pUG fold, a left-handed quadruplex structure that incorporates 12 repeats, but longer pUG tails are more active. Here, we investigate the structure, folding kinetics and dynamics of long pUG RNAs. RNAs with 24 or more repeats slowly form compact, double pUG folds. The forward rate of pUG fold formation in vitro is length-dependent with a half-life (t1/2) of 13 minutes, while the unfolding rate is very slow (t1/2 ~5 days). Long pUG RNAs display biphasic dynamics with an additional, faster unfolding phase (t1/2 ~30 min). The amplitude of the faster phase indicates partial unfolding. From these data we propose a dynamic model for segmental register exchange and double pUG fold formation in long pUG RNAs. These data broaden our understanding of the structure and dynamics of long pUG RNAs and have implications for understanding the roles of pUG folds in biology and disease.