Spatial epigenomic niches underlie glioblastoma cell state plasticity
Spatial epigenomic niches underlie glioblastoma cell state plasticity
Kint, S.; Younes, S. T.; Bao, S.; Long, G.; Wouters, D.; Stephenson, E.; Lou, X.; Zhong, M.; Zhang, D.; Su, G.; Enninful, A.; Yang, M.; Chen, H.-M.; Ellestad, K.; Anderson, C.; Moliterno, J.; Kelly, J.; Chan, J. A.; Sifrim, A.; Zhou, J.; Nikolic, A.; Fan, R.; Gallo, M.
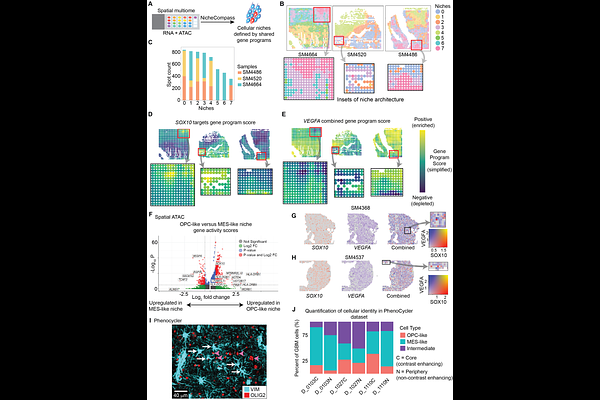
AbstractIDH-wildtype glioblastoma (GBM) is an aggressive brain tumor with poor survival and few therapeutic options. Transcriptionally-defined cell states coexist in GBM and occupy defined regions of the tumor. Evidence indicates that GBM cell states are plastic, but it remains unclear if they are determined by the underlying epigenetic state and/or by microenvironmental factors. Here, we present spatially-resolved epigenomic profiling of human GBM tissues that implicate chromatin structure as a key enabler of cell plasticity. We report two epigenetically-defined and spatially-nested tumor niches. Each niche activates short-range molecular signals to maintain its own state and, surprisingly, long-range signals to reinforce the state of the neighboring niche. The position of a cell along this gradient-like system of opposing signals determines its likelihood to be in one state or the other. Our results reveal an intrinsic system for cell plasticity that is encoded in the chromatin profiles of two adjacent niches that dot the topological architecture of GBM in cartesian space.