Extending the Gaussian Network Model: Integrating Local, Allosteric, and Structural Factors for Improved Residue-Residue Correlation Analysis
Extending the Gaussian Network Model: Integrating Local, Allosteric, and Structural Factors for Improved Residue-Residue Correlation Analysis
Erman, B.
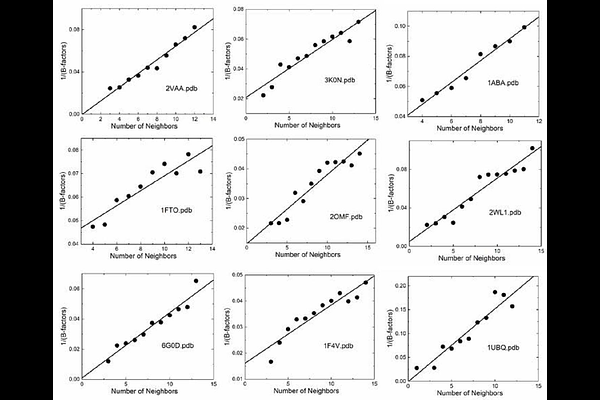
AbstractThe Gaussian Network Model (GNM) has been successful in explaining protein dynamics by modeling proteins as elastic networks of alpha carbons connected by harmonic springs. However, its uniform interaction assumption and neglect of higher-order correlations limit its accuracy in predicting experimental B-factors and residue cross-correlations critical for understanding allostery and information transfer. This study introduces an information-theoretic enhancement to the GNM, incorporating mutual information-based corrections to the Kirchhoff matrix to account for multi-body interactions and contextual residue dynamics. By iteratively optimizing B-factor predictions and applying a Monte Carlo-driven maximum entropy approach to refine covariances, our method achieves significant improvements, reducing RMSDs between predicted and experimental B-factors by 26-46% across eight representative proteins. The model contextualizes residue assignments based on local density, secondary structure, solvent exposure, and allosteric roles, showing complex dynamic patterns beyond simple neighbor counts. Enhanced predictions of mutual information and entropy transfer in proteins like KRAS highlight improved capture of allosteric communication pathways. This evolvable framework, capable of incorporating additional effects and utilizing contextual residue assignments, enables precise studies of mutation effects on protein dynamics, with improved cross-correlation predictions potentially increasing accuracy in drug design and function prediction.