Transposable element products, functions, and regulatory networks in Arabidopsis
Transposable element products, functions, and regulatory networks in Arabidopsis
Borreda, C.; Leduque, B.; Colot, V.; Quadrana, L.
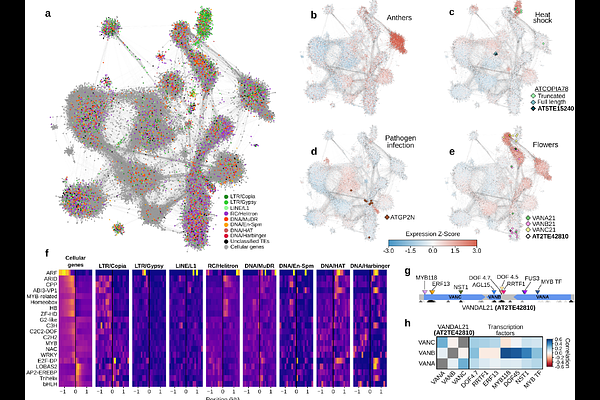
AbstractTransposable elements (TEs) are DNA sequences with the ability to propagate themselves within genomes. Their mobilization is catalyzed by self-encoded factors, yet these factors have been poorly investigated. Here, we leveraged extensive long- and short-read transcriptome data, structural predictions, and regulatory networks analyses, to construct a comprehensive atlas of TE transcripts and their encoded products in the model organism Arabidopsis thaliana. We uncovered hundreds of transcriptionally competent TEs, each potentially encoding multiple proteins either through distinct genes, alternative splicing, or post-translational processing. Structural-based protein analyses revealed hitherto unidentified domains, uncovering proteins with multimerization and DNA binding domains forming macromolecular complexes. Furthermore, we demonstrate that TE expression is highly intertwined with the transcriptional network of cellular genes, and identified transcription factors and cis-regulatory elements associated with their coordinated expression during development or in response to environmental cues. This comprehensive functional atlas provides a valuable resource for studying the mechanisms involved in transposition and their consequences for genome and organismal function.