Benchmarking Genomic Variant Calling Tools in Inbred Mouse Strains: Recommendations and Considerations
Benchmarking Genomic Variant Calling Tools in Inbred Mouse Strains: Recommendations and Considerations
Garretson, A. C.; Blanco-Berdugo, L.; Roberts, A.; Dumont, B. L.
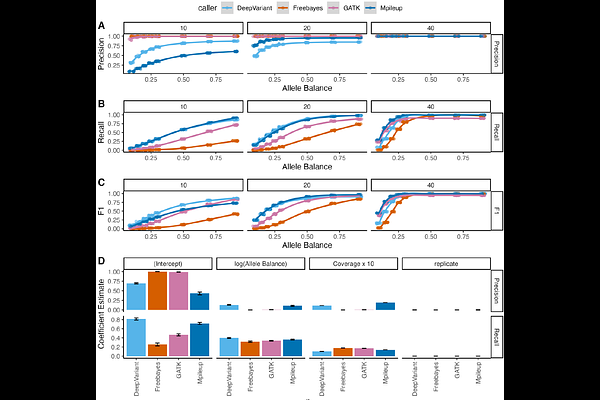
AbstractWith the growing affordability of whole genome sequencing, variant identification has become an increasingly common task, but there are many challenges due to both technical and biological factors. In recent years, the number of software packages available for variant calling has rapidly increased. Understanding the benefits and drawbacks of different tools is important in setting leading practices and highlighting limitations. These considerations are crucial in model organism research, as many variant calling programs assume outbred genomes and implicit heterozygosity, which may not apply to inbred laboratory models. Here, we present an analysis of variant calling tools and their performance in the simulated genomes of the C57BL/6J inbred laboratory mouse and nine non-reference laboratory strains. Our findings reveal a tradeoff between the recall and precision of tools. Balancing these considerations, we show that an optimal call set is obtained by using an ensemble approach, but specific variant calling recommendations vary by strain and analytical goals. Further, we highlight filters improving the performance of different variant calling tools, both for the discovery of rare variants and in the discovery of strain polymorphisms. In summary, our work provides best practices for calling and filtering genomic variants in inbred organisms, particularly laboratory mice.