Two de novo transcriptome assemblies and functional annotations from juvenile cuttlefish (Sepia officinalis) under various metal and pCO2 exposure conditions
Two de novo transcriptome assemblies and functional annotations from juvenile cuttlefish (Sepia officinalis) under various metal and pCO2 exposure conditions
Sol Dourdin, T.; Minet, A.; Pante, E.; Lacoue-Labarthe, T.
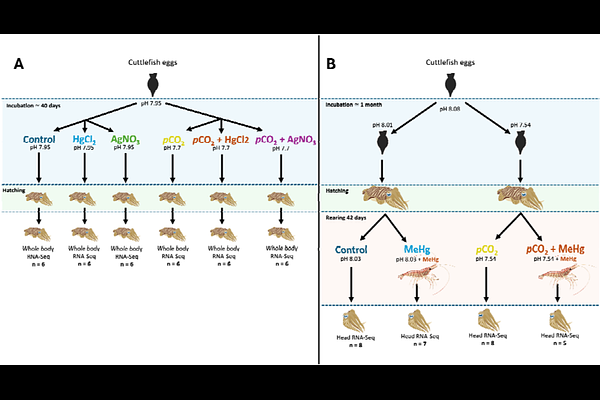
AbstractThe cuttlefish Sepia officinalis is a precious model in behavioural and neurobiology studies. It is currently facing combined environmental changes related to the anthropogenic global change. However, genomic resources available to support investigations tackling this issue are still scarce. Therefore, we present two annotated de novo transcriptome assemblies from recently hatched (whole body) and one-month-old (head) Sepia officinalis juveniles. Both assemblies rely on an important read depth validated by a pseudo-rarefaction analysis, and gathered several individuals from various metal and pCO2 exposure conditions. After redundancy reduction, assemblies from newly hatched and one-month-old individuals comprised 244,772 and 397,569 transcripts with 39,804 and 49,402 putative ORFs, respectively, and an average annotation rate of 60%. Assemblies were compared to each other, revealing age-specific transcriptomic landscapes. These two assemblies constitute highly valuable genomic resources facilitating the investigation of transcriptomic endpoints in environmental studies considering coleoid cephalopods.