Resolving Oligomeric States of Photoactivatable Proteins in Live Cells via Photon Counting Histogram Analysis
Resolving Oligomeric States of Photoactivatable Proteins in Live Cells via Photon Counting Histogram Analysis
Camp, T.; Li, Z.; Li, Y.; Oh, T.-J.; Zhang, K.
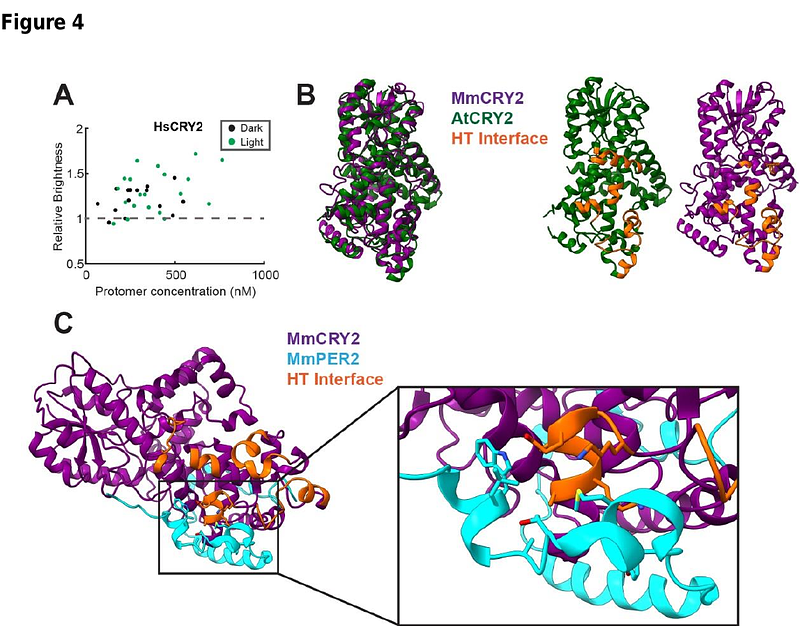
AbstractOligomerization of photoactivatable proteins is widely used in optogenetics to modulate protein activity and regulate biological processes. However, their oligomerization states remain challenging to quantify in living cells. We applied photon counting histogram (PCH) analysis to quantify the oligomerization of two commonly used photoactivatable proteins, Vaucheria frigida Aureochrome light-oxygen-voltage (VfAuLOV) and Arabidopsis Thaliana cryptochrome 2 (AtCRY2), under dark and light conditions, providing a direct measurement of oligomerization states in live cells. Under blue light stimulation, VfAuLOV primarily forms dimers, whereas AtCRY2 forms higher order multimers in HEK293T cells. At intracellular concentrations above 1000 nM, AtCRY2 transitions into tetramers upon light stimulation, consistent with structural data obtained from cryo-electron microscopy. Unexpectedly, AtCRY2 (W374), a constitutively active mutant, still exhibits light-induced oligomerization. Human cryptochrome 2, in contrast, shows light-independent oligomerization at the intracellular concentration above 100 nM, below which the monomeric state dominates. Optogenetic signaling outcomes, demonstrated by light-induced lytic cell death, align with the oligomerization state of tested proteins. This study presents a quantitative framework for elucidating protein oligomerization in live cells, thereby enhancing the understanding of optogenetic mechanisms and their role in cellular signaling.