Structural basis for TORC2 activation
Structural basis for TORC2 activation
Zou, L.; Tettamanti, M. G.; Bergmann, A.; Loewith, R.; Tafur, L.
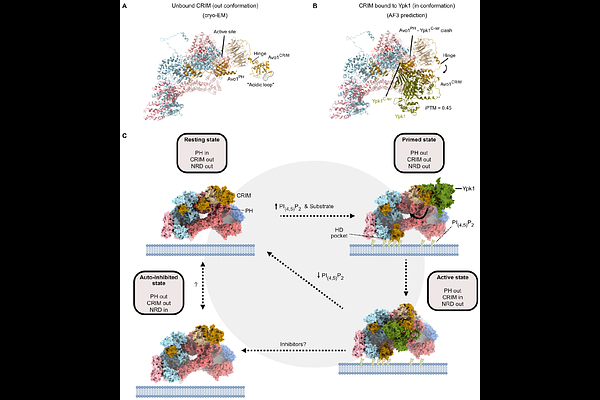
AbstractThe Target of Rapamycin Complex 2 (TORC2) is a central node in signaling feedback loops serving to maintain biophysical homeostasis of the plasma membrane (PM). How TORC2 is regulated by mechanical perturbation of the PM is not well understood. To address this, we determined the cryo-electron microscopy structure of endogenous yeast TORC2 to up to 2.2 A resolution. Our model refines the position and interactions of TORC2-specific subunits, providing a structural basis for the differential assembly of Tor2 into TORC2. Furthermore, we observe the insertion of the pleckstrin-homology domain of the Avo1 subunit into the Tor2 active site, providing a regulatory mechanism by phosphoinositides. Structure-guided functional experiments reveal a potential TORC2 membrane binding surface and a positively charged pocket in the Avo3 subunit that is necessary for TORC2 activation. Collectively, our data suggest that signaling phosphoinositides activate TORC2 by membrane-induced structural rearrangements via concerted action of conserved regulatory subunits.