TRPM8 protein dynamics correlates with ligand structure and cellular function
TRPM8 protein dynamics correlates with ligand structure and cellular function
Mebrat, M. D.; Luu, D. D.; Hilton, J. K.; Kim, M.; Parrott, K.; Cherry, B. R.; Levitus, M.; Journigan, V. B.; Van Horn, W. D.
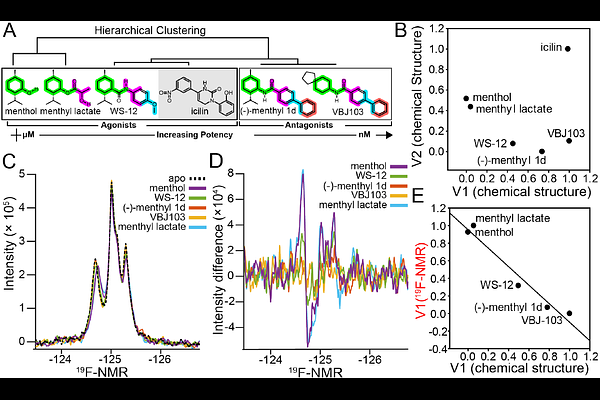
AbstractProtein dynamics have emerged as a key feature associated with function in various systems. Here, NMR-based studies coupled with computational cheminformatics and cellular function are leveraged to identify a relationship between the human cold and menthol receptor TRPM8 dynamics, chemical structure, and cellular potency. TRPM8 is a validated target for a variety of pain indications but generally has been clinically limited by on-target side effects impacting thermosensing and thermoregulation. This study shows that cheminformatic analysis of a TRPM8 regulating small molecule ligand library correlates with cellular function. Electrophysiology studies further validate the relationship and show a correlation between chemical structure and functional features such as compound potency. Solution NMR studies of the TRPM8 voltage sensing-like domain, which houses the canonical menthol ligand binding site, show that ligand binding conformationally selects NMR-detected TRPM8 dynamics in a manner that quantitatively correlates with chemical structure. The relationship between chemical structure and protein dynamics can be used predictively, where a chemical structure is predictive of dynamics in a latent reduced dimensionality space. Moreover, the robustness of the conformational selection of the dynamic ensemble is evaluated by varying related and divergent chemotypes, signal-to-noise sensitivity, and sample bias. Taken together, this study identifies that protein dynamics can serve as a quantifiable bridge between chemical structure and cellular function, which has implications for drug discovery in difficult systems.