Genomic insights into the virulence repertoire and hemibiotrophic lifestyle of the grapevine black rot pathogen Phyllosticta ampelicida
Genomic insights into the virulence repertoire and hemibiotrophic lifestyle of the grapevine black rot pathogen Phyllosticta ampelicida
Colombo, M.; Bettinelli, P.; Garcia, J.; Maddalena, G.; Toffolatti, S. L.; Hausmann, L.; Vezzulli, S.; Masiero, S.; Cantu, D.
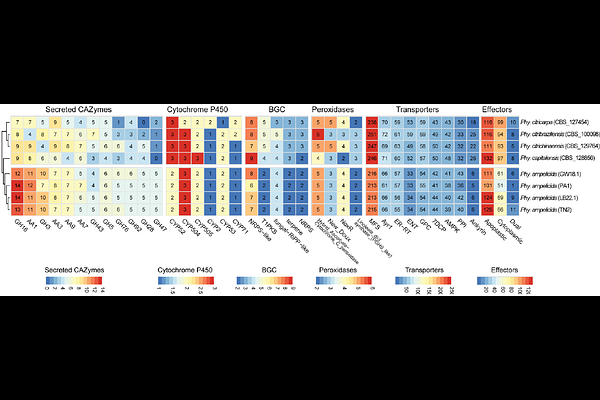
AbstractPhyllosticta ampelicida, the causal agent of grapevine black rot, is a globally emerging pathogen that infects all grapevine green tissues, with young shoots and berries being particularly susceptible. Severe infections can result in total crop loss. To investigate its virulence repertoire, we generated a high-quality genome assembly of strain GW18.1 using long-read sequencing, resulting in 22 scaffolds, including four complete chromosomes and 12 chromosome arms, with a total genome size of 35.6 Mb and 10,289 predicted protein-coding genes. Two additional strains (TN2 and LB22.1) were sequenced with short reads to assess intraspecies diversity. Comparative genomics revealed a conserved virulence factor repertoire, including 314 carbohydrate-active enzymes (CAZymes), 17 cytochrome P450s, 35 peroxidases, and 20 secondary metabolite biosynthetic gene clusters (BGCs). Trophic lifestyle prediction based on gene content supports a biotrophic-like lifestyle consistent with hemibiotrophic pathogens. Broader comparisons with other Phyllosticta species and ten plant-pathogenic fungi pointed to species-specific features, while analysis of gene family evolution identified expansions and contractions in transporters and CAZymes. These genomic resources will support efforts to better understand and manage grapevine black rot. .