Characterization of a potential antibiotic target site on the ribosome
Characterization of a potential antibiotic target site on the ribosome
Golstein, M.; Wang, Y.; Byju, S.; Mohanty, U.; Whitford, P. C.
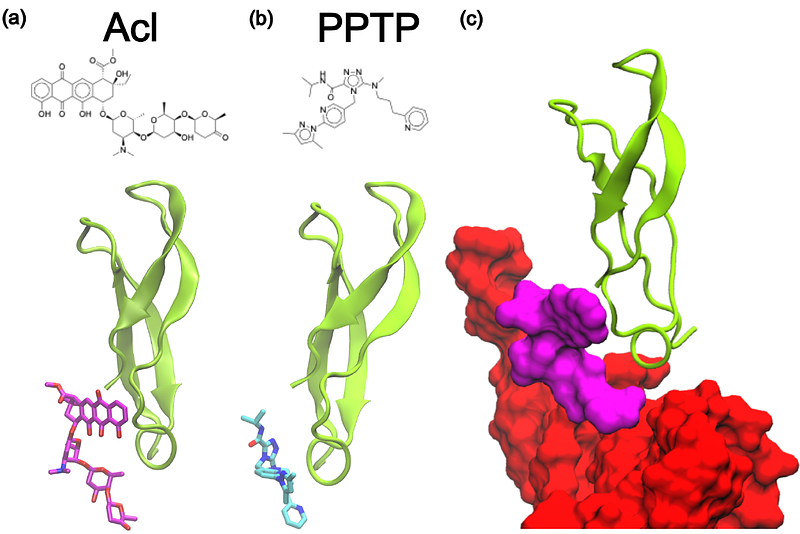
AbstractThe ribosome is a known antibiotic target, where various classes of small molecules impact different stages of translation. There are multiple modes of function, such as interfering with the decoding process, impeding movement of the nascent protein chain and occluding the catalytic center. In the present study, we used a range of computational methods to demonstrate a new mechanism by which small molecules may impede translation in the bacterial ribosome. Specifically, we use a computational screen to identify small molecules that can bind the ribosomal protein L33 in a region that is generally conserved in bacterial species. In addition, the binding position allows it to introduce a steric obstacle that impedes the P/E hybrid-state formation steps. Using molecular dynamics simulations, we show how binding to L33 can slow down the kinetics of tRNA molecules on the ribosome. Since L33 is not present in the cytosolic human ribosome and has a distinct sequence in the human mitochondrial ribosome, this binding site may serve as a novel target for future antibiotic and antimicrobial design efforts. This analysis provides insights into how to optimize the inhibitory effects of L33-targeting molecules, in order to develop a new class of broad-spectrum antibiotics/antimicrobials.