A genome-wide approach uncovers the suite of genes important for swarming motility in the biocontrol bacteria Pseudomonas protegens Pf-5
A genome-wide approach uncovers the suite of genes important for swarming motility in the biocontrol bacteria Pseudomonas protegens Pf-5
Fabian, B. K.; Foster, C.; Asher, A.; Elbourne, L. D.; Hassan, K. A.; Tetu, S. G.; Paulsen, I. T.
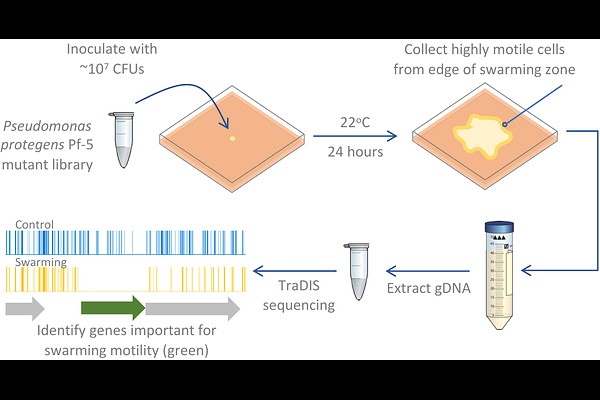
AbstractMotility is a crucial bacterial trait, important for competitive success in many contexts. Plant growth promoting rhizobacteria, such as Pseudomonas protegens Pf-5, can protect agricultural crops against disease, relying on motility to access and colonize the plant rhizosphere. Swarming motility is the multicellular movement of bacteria powered by rotating flagella across a semi-solid surface which allows bacteria to move rapidly, search for and acquire food, avoid toxic substances, and colonize new niches. Here we performed genome-wide investigations, using transposon-directed insertion site sequencing (TraDIS) to identify 136 genes involved in P. protegens Pf-5 swarming motility. This method identified flagella biosynthesis, assembly, structure and regulatory genes, as expected. We also identified an additional suite of genes important for swarming, including chemotaxis, c-di-GMP turnover, cell division, peptidoglycan turnover, lipopolysaccharide biosynthesis, metabolism of thiamine (vitamin B1) and transport of pyoverdine. Overall, this study increases our understanding of pseudomonad motility and expands the breadth of lifestyles and habitats examined in relation to swarming motility. Understanding the genetic basis of swarming motility in biocontrol bacteria is important for successful application of these important plant growth promoting microbes in agricultural settings.