The planktonic microbiome of the Great Barrier Reef
The planktonic microbiome of the Great Barrier Reef
Robbins, S. J.; Dougan, K.; Terzin, M.; Zaugg, J.; Bell, S. C.; Laffy, P. W.; Engelberts, J. P.; Webster, N. S.; Hugenholtz, P.; Bourne, D. G.; Yeoh, Y. K.
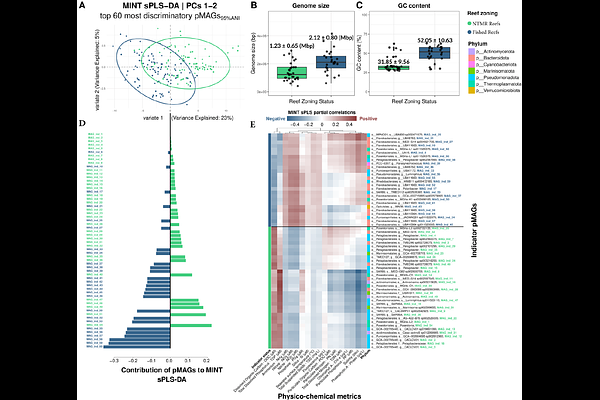
AbstractLarge genome databases have markedly improved our understanding of marine microorganisms. Although these resources have focused on prokaryotes, genomes from many dominant marine lineages, such as Pelagibacter and Prochlorococcus, are conspicuously underrepresented. Here, we present the Great Barrier Reef Microbial Genomes Database (GBR-MGD) comprising 5,283 prokaryotic genomes obtained from GBR seawater samples using Nanopore sequencing, including a collection of high quality genomes of underrepresented groups. We show that standard short read assemblies miss these populations due to a combination of strain heterogeneity and low GC% sequencing bias. The GBR-MGD also comprises 19 chromosome-level picoeukaryote and 808,585 viral genomes, including a newly described clade of marine Crassvirales. We demonstrate the use of the GBR-MGD to identify indicator taxa that can reliably predict the effects of reef management practices, such as the establishment of marine protected zones.