Towards Unraveling Biomolecular Conformational Landscapes with a Generative Foundation Model
Towards Unraveling Biomolecular Conformational Landscapes with a Generative Foundation Model
The OpenComplex Team, ; Ye, Q.
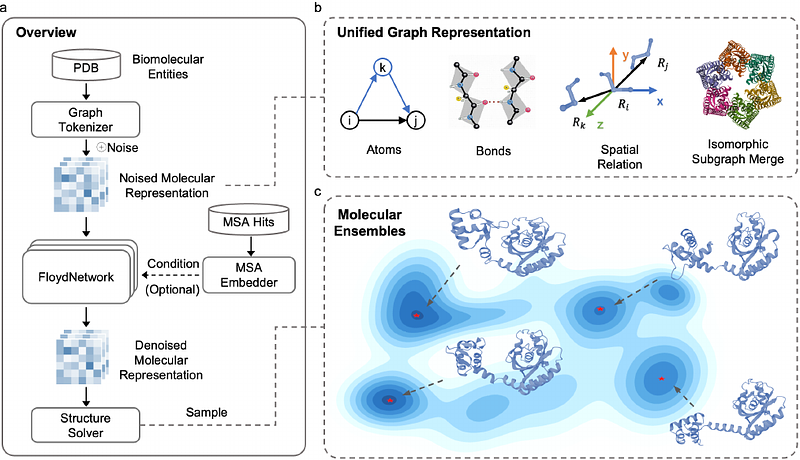
AbstractRecent advances in deep learning have revolutionized biomolecular structure prediction, yet a significant challenge remains: biological function emerges from dynamic equilibria across multiple conformational states rather than a single molecular structure. Here, we present OpenComplex2 (OC2), a generative model that efficiently sample biomolecular conformational ensembles. OC2 integrates a unified graph representation capturing diverse molecular inputs with diffusion-based sampling through our FloydNetwork architecture, which operates simultaneously across atomic, residue, and motif hierarchies. This multi-level approach balances fine-grained precision with computational efficiency, enabling conformational landscape sampling in hours rather than the weeks required by traditional simulations. Through comprehensive evaluation, we demonstrate that OC2 accurately reproduces experimentally determined protein and RNA ensembles, generates conformational distributions comparable to millisecond-scale molecular dynamics simulations, and captures functionally relevant transitions in important biological scenarios including allostery, induced-fit mechanisms, and cryptic pocket formation. OC2 also effectively models small molecule conformations, maintains competitive accuracy on structure prediction benchmarks, and scales to ultra-large molecular assemblies exceeding 15,000 residues. By bridging static structure prediction and dynamic ensemble characterization, OC2 advances our understanding of how biomolecular dynamics give rise to biological function.