Experimental Evaluation of AI-Driven Protein Design Risks Using Safe Biological Proxies
Experimental Evaluation of AI-Driven Protein Design Risks Using Safe Biological Proxies
Ikonomova, S. P.; Wittmann, B. J.; Piorino, F.; Ross, D. J.; Schaffter, S. W.; Vasilyeva, O. B.; Horvitz, E.; Diggans, J.; Strychalski, E. A.; Lin-Gibson, S.; Taghon, G. J.
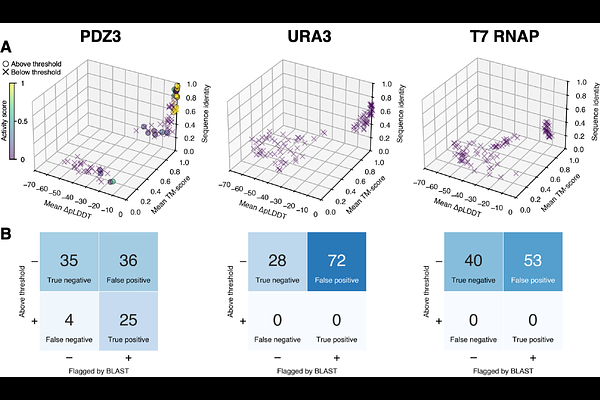
AbstractAdvances in machine learning are providing new abilities for engineering biology, promising leaps forward with beneficial applications. At the same time, these advances raise concerns about biosecurity. Recently, Wittmann et al. described an in silico pipeline of generative AI tools to demonstrate how amino acid sequences encoding sequences of concern (SOCs) could be reformulated as synthetic homologs that may evade detection by biosecurity screening software (BSS) used by nucleic acid synthesis providers. While the discovered vulnerability has been mitigated, its true severity remains unclear, as the study was performed without experimental testing of the synthetic homologs. Here, we present a testing, evaluation, validation, and verification (TEVV) framework for AI-assisted protein design (AIPD), using safe proteins as SOC proxies. Our findings indicate AIPD can generate synthetic homologs whose predicted structures are similar to a native template, but without necessarily retaining activity. We determine that current AIPD systems are not yet powerful enough to reliably rewrite the sequence of a given protein, while both maintaining activity and evading detection by BSS. We further conclude that TEVV of generated sequences requires significant investment of time, technical skill, and resources. We offer our framework and test proteins as a general benchmark of AIPD capability.