Integrative Transcriptomic and miRNA Analysis Reveals Immune Suppression and Metabolic Reprogramming in FGFR3-TACC3 Fusion-Positive versus Fusion-Negative Bladder Cancer
Integrative Transcriptomic and miRNA Analysis Reveals Immune Suppression and Metabolic Reprogramming in FGFR3-TACC3 Fusion-Positive versus Fusion-Negative Bladder Cancer
Mishra, D.; Agrawal, S.; Malik, D.; Pathak, E.; Mishra, R.
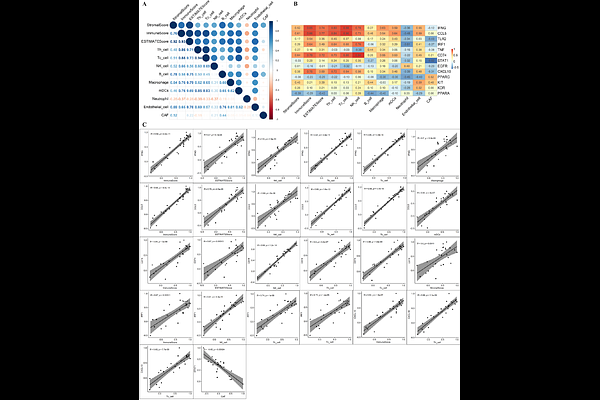
AbstractFGFR3-TACC3 gene fusion drives a molecularly distinct subset of bladder cancer characterized by post-transcriptional immune evasion and metabolic rewiring. To elucidate the transcriptomic and regulatory consequences of this fusion, we performed an integrative analysis of mRNA and miRNA expression profiles in fusion-positive versus fusion-negative bladder tumors using TCGA-derived RNA-Seq and miRNA-Seq data. Fusion-positive tumors exhibited upregulation of mitochondrial oxidative phosphorylation and ribosomal genes (e.g., COX5B, ATP5F1D, RPS15) and marked downregulation of immune regulatory genes (e.g., CD4, STAT3, IL6, CD86, PTPRC). Pathway enrichment revealed suppression of JAK/STAT, PD-L1 signaling, and cytokine networks, consistent with a fusion-driven immunosuppressive phenotype. We identified fusion-specific miRNA-mRNA regulatory interactions, including upregulated miRNAs (miR-1226-3p, miR-149-5p, miR-301b-3p) that target immune transcripts and downregulated miRNAs (miR-199b-5p, miR-214-3p) that normally suppress mitochondrial and ribosomal genes. In contrast, fusion-negative tumors displayed an immunoreactive signature marked by elevated IFNG, CXCL10, and IRF1 expression and downregulation of immune-suppressive miRNAs (miR-125a-5p, miR-23b-3p). Minimum free energy analysis and AlphaFold3-based AGO2-RISC docking confirmed structural feasibility of 3D duplex formation for 13 miRNA-target pairs, supporting active silencing complexes. Immune deconvolution revealed reduced infiltration of cytotoxic T cells, NK cells, and dendritic cells in fusion-positive tumors, while fusion-negative tumors showed robust immune cell presence. Key gene-immune cell correlations further confirmed differential immune architecture: SRC and CD4 were strongly linked to suppressed infiltration in fusion-positive tumors, while IFNG and CXCL10 positively correlated with T and NK cell abundance in fusion-negative cases. ROC analysis highlighted COMT, RPS15, and miR-187-3p as robust classifiers of fusion-positive tumors (AUC > 0.9), and CXCL10, CD74, and miR-142 for fusion-negative tumors. Connectivity Map analysis highlighted Lofexidine and Lenalidomide as candidate compounds predicted to reverse fusion-specific transcriptional programs. Collectively, our findings define immune suppression and metabolic reprogramming as core features of FGFR3-TACC3 fusion-positive bladder cancer and nominate fusion-specific miRNA-mRNA interactions as candidate biomarkers and therapeutic targets.